In this tutorial, you will be introduced to the command line. We have selected a set of commands we think will be useful in general to a wide range of audience. We have created a RStudio Cloud Project to ensure that all readers are using the same environment while going through the tutorial. Our goal was to ensure that after completing this tutorial, readers should be able to use the shell for version control, managing cloud services (like deploying your own shiny server etc.), execute commands in R & RMarkdown and execute R scripts in the shell. Apart from learning shell commands, the tutorial will also focus on
- exploring R release names
- mapping shell commands to R functions
- RStudio Terminal
- executing shell commands from R using
system2()orprocessx::run() - execute shell commands in RMarkdown
- execute R scripts in the shell
If you want a deeper understanding of using command line for data science, we suggest you read Data Science at the Command Line. Software Carpentry too has a lesson on shell. We have listed more references at the end of the tutorial for the benefit of the readers.
Table of Contents
Resources
Below are the links to all the resources related to this post:
You can try our free online course Command Line Basics for R Users if you prefer to learn through self paced online courses or our ebook if you like to read the tutorial in a book format.
Introduction
What is Shell/Terminal?
Shell is a text based application for viewing, handling & manipulating files. It takes in commands and passes them on to the operating system. It is also known as
- CLI (Command Line Interface)
- Bash (Bourne Again Shell)
- Terminal
It is sufficient to know a handful of commands to get started with the shell.
Launch Terminal
Although we will use the terminal in RStudio on RStudio Cloud, we should still know how to launch the terminal in different operating systems.
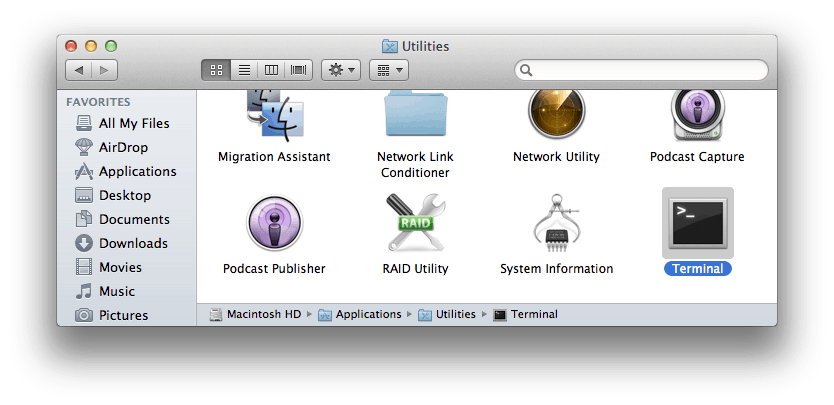
mac
Applications -> Utility -> Terminal
Windows
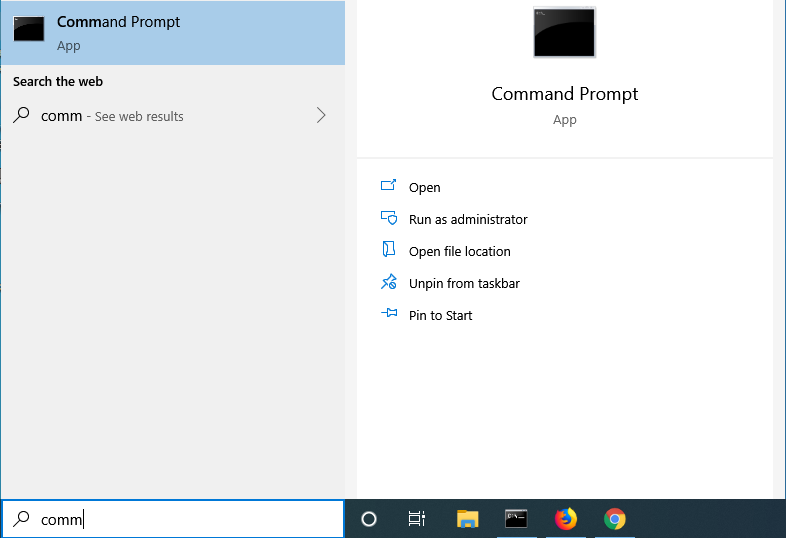
Option 1
Go to the Start Menu or screen and enter Command Prompt in the search field.
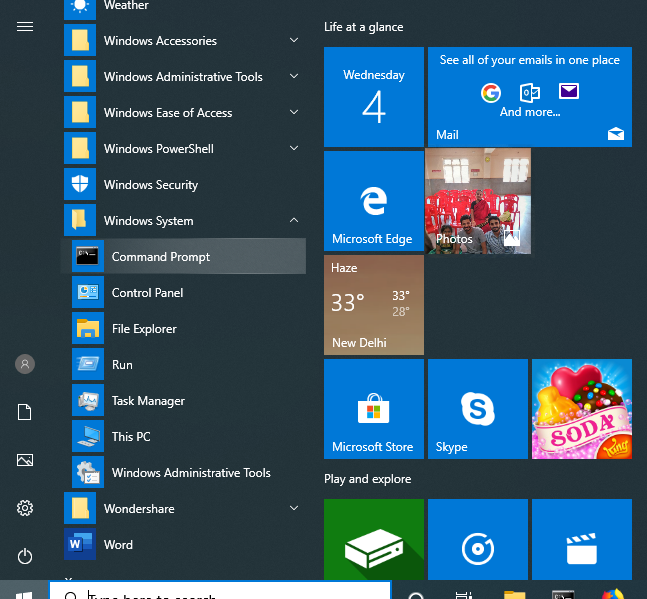
Option 2
Start Menu -> Windows System -> Command Prompt
Option 3
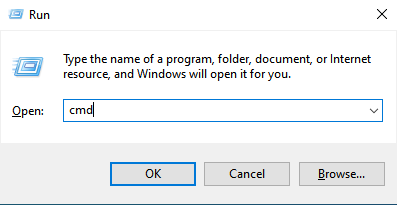
Hold the Windows key and press the R key to get a Run window. Type cmd in the box and click on the OK button.
Linux
- Applications -> Accessories -> Terminal
- Applications -> System -> Terminal
Windows Subsystem for Linux
If you want to use bash on Windows, try the Windows subsystem for Linux. It only works on 64 bit Windows 10. Below are the steps to enable Windows subsystem fro Linux:
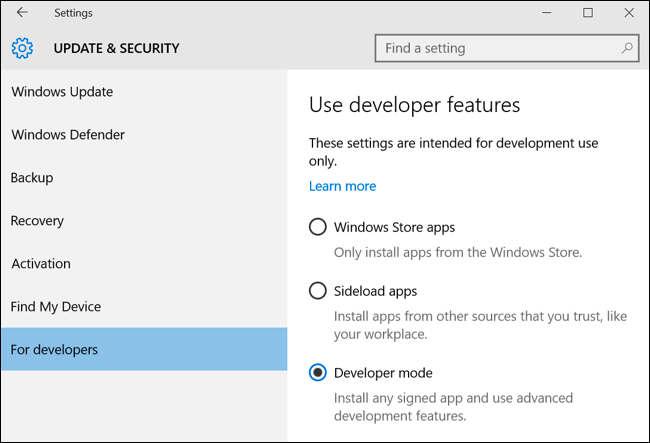
Step 1 - Enable Developer Mode
To enable Developer Mode open the Settings app and head to Update & Security > For Developers. Press the Developer Mode switch.
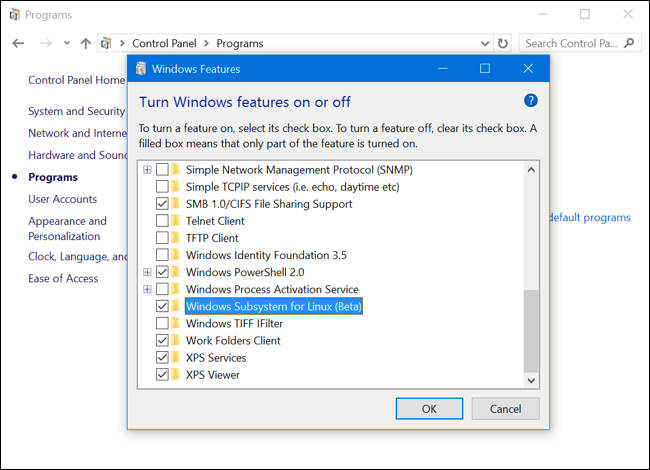
Step 2 - Enable Windows Subsystem for Linux
To enable the Windows Subsystem for Linux (Beta), open the Control Panel, click Programs and Features, and click Turn Windows Features On or Off in left side bar under Programs and Features. Enable the Windows Subsystem for Linux (Beta) option in the list here and click OK. After you do, you’ll be prompted to reboot your computer. Click Restart Now to reboot your computer and Windows 10 will install the new feature.
Step 3 - Install your Linux Distribution of Choice
Open the Microsoft store and choose your favorite Linux distribution.
In the distro’s page, click on “Get”.
Launch the distro from the Start Menu.
You can learn more about the Windows Subsystem for Linux here.
RStudio Terminal
RStudio introduced the terminal with version 1.1.383. The terminal tab is next to the console tab. If it is not visible, use any of the below methods to launch it
- Shift + Alt + T
- Tools -> Terminal -> New Terminal
Note, the terminal depends on the underlying operating system. To learn more about the RStudio terminal, read this article or watch this webinar. In this book, we will use the RStudio terminal on RStudio Cloud to ensure that all users have access to Linux bash. You can try all the commands used in this book on your local system as well except in case of Windows users.
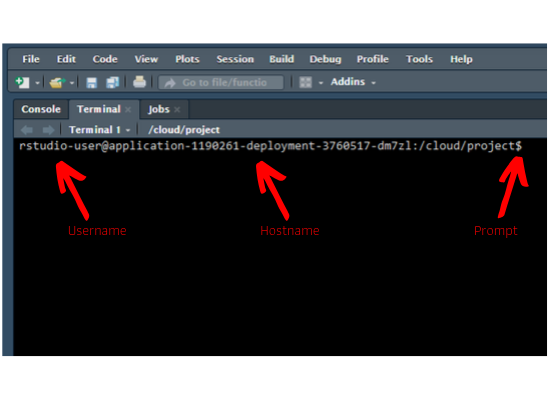
Prompt
As soon as you launch the terminal, you will see the hostname, machine name and
the prompt. In case of mac & Linux users, the prompt is $. For Windows users, it is >.
| OS | Prompt |
|---|---|
| macOS | $ |
| Linux | $ |
| Windows | > |
Get Started
To begin with, let us learn to display
- basic information about the user
- the current date & time
- the calendar
- and clear the screen.
| Command | Description |
|---|---|
whoami
|
Who is the user? |
date
|
Get date, time and timezone |
cal
|
Display calendar |
clear
|
Clear the screen |
whoami prints the effective user id i.e. the name of the user who runs the command. Use it to verify the user as which you are logged into the system.
whoami## aravinddate will display or change the value of the system’s time and date information.
date## Wed Jun 10 22:45:47 IST 2020cal will display a formatted calendar and clear will clear all text on the screen and display a new prompt. You can clear the screen by pressing Ctrl + L as well.
calIn R, we can get the user information from Sys.info() or whoami() from the whoami package. The current date & time are returned by Sys.date() & Sys.time(). To clear the R console, we use Ctrl + L.
| Command | R |
|---|---|
whoami
|
Sys.info() / whoami::whoami()
|
date
|
Sys.date() / Sys.time()
|
cal
|
|
clear
|
Ctrl + L
|
Help/Documentation
Before we proceed further, let us learn to view the documentation/manual pages of the commands.
| Command | Description |
|---|---|
man
|
Display manual pages for a command |
whatis
|
Single line description of a command |
man is used to view the system’s reference manual. Let us use it to view the documentation of the whatis command which we will use next.
man whatis## WHATIS(1) Manual pager utils WHATIS(1)
##
## NAME
## whatis - display one-line manual page descriptions
##
## SYNOPSIS
## whatis [-dlv?V] [-r|-w] [-s list] [-m system[,...]] [-M path] [-L
## locale] [-C file] name ...
##
## DESCRIPTION
## Each manual page has a short description available within it. whatis
## searches the manual page names and displays the manual page descripâ€
## tions of any name matched.
##
## name may contain wildcards (-w) or be a regular expression (-r). Using
## these options, it may be necessary to quote the name or escape (\) the
## special characters to stop the shell from interpreting them.
##
## index databases are used during the search, and are updated by the
## mandb program. Depending on your installation, this may be run by a
## periodic cron job, or may need to be run manually after new manual
## pages have been installed. To produce an old style text whatis dataâ€
## base from the relative index database, issue the command:
##
## whatis -M manpath -w '*' | sort > manpath/whatis
##
## where manpath is a manual page hierarchy such as /usr/man.
##
## OPTIONS
## -d, --debug
## Print debugging information.
##
## -v, --verbose
## Print verbose warning messages.
##
## -r, --regex
## Interpret each name as a regular expression. If a name matches
## any part of a page name, a match will be made. This option
## causes whatis to be somewhat slower due to the nature of dataâ€
## base searches.
##
## -w, --wildcard
## Interpret each name as a pattern containing shell style wildâ€
## cards. For a match to be made, an expanded name must match the
## entire page name. This option causes whatis to be somewhat
## slower due to the nature of database searches.
##
## -l, --long
## Do not trim output to the terminal width. Normally, output will
## be truncated to the terminal width to avoid ugly results from
## poorly-written NAME sections.
##
## -s list, --sections list, --section list
## Search only the given manual sections. list is a colon- or
## comma-separated list of sections. If an entry in list is a simâ€
## ple section, for example "3", then the displayed list of
## descriptions will include pages in sections "3", "3perl", "3x",
## and so on; while if an entry in list has an extension, for examâ€
## ple "3perl", then the list will only include pages in that exact
## part of the manual section.
##
## -m system[,...], --systems=system[,...]
## If this system has access to other operating system's manual
## page names, they can be accessed using this option. To search
## NewOS's manual page names, use the option -m NewOS.
##
## The system specified can be a combination of comma delimited
## operating system names. To include a search of the native operâ€
## ating system's manual page names, include the system name man in
## the argument string. This option will override the $SYSTEM
## environment variable.
##
## -M path, --manpath=path
## Specify an alternate set of colon-delimited manual page hierarâ€
## chies to search. By default, whatis uses the $MANPATH environâ€
## ment variable, unless it is empty or unset, in which case it
## will determine an appropriate manpath based on your $PATH enviâ€
## ronment variable. This option overrides the contents of $MANâ€
## PATH.
##
## -L locale, --locale=locale
## whatis will normally determine your current locale by a call to
## the C function setlocale(3) which interrogates various environâ€
## ment variables, possibly including $LC_MESSAGES and $LAwhatis displays short manual page descriptions (each manual page has a short description available within it).
whatis ls## ls (1) - list directory contentsYou will find tldr.sh very useful while exploring new commands and there is a related R package, tldrrr as well.
# devtools::install_github("kirillseva/tldrrr")
tldrrr::tldr("pwd")## pwd
##
## Print name of current/working directory.
##
## • Print the current directory:
##
## pwd
##
## • Print the current directory, and resolve all symlinks (i.e. show the "physical" path):
##
## pwd -PFile Management
In this section, we will explore commands for file management including:
- create new file/change timestamps
- copying files
- renaming/moving files
- deleting files
- comparing files
| Command | Description |
|---|---|
touch
|
Create empty file(s)/change timestamp |
cp
|
Copy files & folders |
mv
|
Rename/move file |
rm
|
Remove/delete file |
diff
|
Compare files |
Create new file
touch modifies file timestamps which is information associated with file modification. It can be any of the following:
- access time (the last time the file was read)
- modification time (the last time the contents of the file was changed)
- change time (the last time the file’s metadata was changed)
If the file does not exist, it will create an empty file of the same name. Let us use touch to create a new file myanalysis.R.
touch myanalysis.R
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myanalysis.R
## myfiles
## mypackage
## myproject
## myproject1
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipCopy Files/Folders
cp makes copies of files and directories. The general form of the command is
cp source destination. By default, it will overwrite files without prompting for confirmation so be cautious while copying files or folders.
Copy files in same folder
Let us create a copy of release_names.txt file and name it as release_names_2.txt.
cp release_names.txt release_names_2.txt
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myanalysis.R
## myfiles
## mypackage
## myproject
## myproject1
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## release_names_2.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipCopy files into different folder
To copy a file into a different directory/folder, we need to specify the name of the destination folder. If the copied file should have a different name, then we need to specify the new name of the file as well. Let us copy the release_names.txt file into the r_releases folder (we will retain the same name for the file as we are copying it into a different folder).
cp release_names.txt r_releases/release_names.txtLet us check if the file has been copied by listing the files in the r_releases folder using ls.
ls r_releases## release_names.txtCopy folders
How about making copies of folders? Use the -r option to copy entire folders. Let us create a copy of the r folder and name it as r2. The -r option stands for --recursive i.e. copy directories recursively.
cp -r r r2
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myanalysis.R
## myfiles
## mypackage
## myproject
## myproject1
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## release_names_2.txt
## release_names_3.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipMove/Rename Files
mv moves and renames files and directories. Using different options, we can ensure
- files are not overwritten
- user is prompted for confirmation before overwriting files
- details of files being moved is displayed
| Command | Description |
|---|---|
mv
|
Move or rename files/directories |
mv -f
|
Do not prompt for confirmation before overwriting files |
mv -i
|
Prompt for confirmation before overwriting files |
mv -n
|
Do not overwrite existing files |
mv -v
|
Move files in verbose mode |
Let us move the release_names_2.txt file to the r_releases folder.
mv release_names_2.txt r_releasesUse ls to verfiy if the file has been moved. As you can see, release_names_2.txt is not present in the current working directory.
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myanalysis.R
## myfiles
## mypackage
## myproject
## myproject1
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## release_names_3.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipLet us check if release_names_2.txt is present in the r_releases folder. Great! We have successfully moved the file into a different folder.
ls r_releases## release_names.txt
## release_names_2.txtMove files in verbose mode
To view the details of the files being moved/renamed, use the -v option. In the below example, we move the release_names_3.txt file into the r_releases folder using mv.
mv -v release_names_3.txt r_releases## renamed 'release_names_3.txt' -> 'r_releases/release_names_3.txt'Do not overwrite existing files
How do we ensure that files are not overwritten without prompting the user first? In the below example, we will try to overwrite the release_names_2.txt in the r_releases folder using mv and see what happens. But first, let us look at the contents of the release_names_2.txt file using the cat command.
We will look into the cat command in more detail in the next chapter but for the time being it is sufficient to know that it prints contents of a file. The file contains release names of different R versions.
cat r_releases/release_names_2.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeIn our current working directory, we will create another file of the same name i.e. release_names_2.txt but its contents are different from the file in the r_releases folder. It contains the string release_names and nothing else. We will now move this file into the r_releases folder but use the option -n to ensure that the file in the r_releases folder is not overwritten. We can confirm this by printing the contents of the file in the r_releases folder.
The echo command is used to print text to the terminal or to write to a file. We will explore it in more detail in the next chapter.
echo "release_names" > release_names_2.txt
mv -n release_names_2.txt r_releases
cat r_releases/release_names_2.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeAs you can observe, the contents of the file in the r_releases folder has not changed. In the next section, we will learn to overwrite the contents using the -f option.
Do not prompt for confirmation before overwriting files
What if we actually intend to overwrite a file and do not want to be prompted for confirming the same. In this case, we can use the -f option which stands for --force i.e. do not prompt before overwriting. Let us first print the contents of the release_names_2.txt file in the r_releases folder.
cat r_releases/release_names_2.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeNow we will create another file of the same name in the current working directory but with different content and use the -f option to overwrite the file in the r_releases folder. You can see that the contents of the file in the r_releases folder has changed.
echo "release_names" > release_names_2.txt
mv -f release_names_2.txt r_releases
cat r_releases/release_names_2.txt## release_namesRemove/Delete Files
The rm command is used to delete/remove files & folders. Using additional options, we can
- remove directories & sub-directories
- forcibly remove directories
- interactively remove multiple files
- display information about files removed/deleted
| Command | Description |
|---|---|
rm
|
Remove files/directories |
rm -r
|
Recursively remove a directory & all its subdirectories |
rm -rf
|
Forcibly remove directory without prompting for confirmation or showing error messages |
rm -i
|
Interactively remove multiple files, with a prompt before every removal |
rm -v
|
Remove files in verbose mode, printing a message for each removed file |
Remove files
Let us use rm to remove the file myanalysis.R (we created it earlier using the touch command).
rm myanalysis.R
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject1
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipRecursive Deletion
How about folders or directories? We can remove a directory and all its contents including sub-directories using the option -r which stands for --recursive and removes directories and their contents recursively. Let us remove the myproject1 folder and all its contents.
rm -r myproject1
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject2
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipForce Removal
Use the -f option which stands for --force to forciby remove directory and all its contents without prompting for confirmation or showing error messages. Let us remove the myproject2 folder and all its contents.
rm -rf myproject2
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipVerbose Mode
Remove files in verbose mode, printing a message for each removed file. This is useful when you want to see the details of the files being removed. In the below example, we will remove all files with .txt extension from the myfiles folder. Instead of specifying the name of each text file, we use the wildcard * along with .txt i.e. any file with the extension .txt will be removed.
cd myfiles
rm -v *.txt## removed 'release_names.txt'
## removed 'release_names_18.txt'
## removed 'release_names_19.txt'Compare Files
diff stands for difference. It is used to compare files line by line and display differences. It also indicates which lines in one file must be changed to make the files identical. Using additional options, we can
- ignore white spaces while comparing files
- show differences sidy by side
- show differences in unified format
- compare directories recursively
- display names of files that differ
| Command | Description |
|---|---|
diff
|
Compare files & directories |
diff -w
|
Compare files; ignoring white spaces |
diff -y
|
Compare files; showing differences side by side |
diff -u
|
Compare files; show differences in unified format |
diff -r
|
Compare directories recursively |
diff -rq
|
Compare directories; show the names of files that differ |
Compare Files
Let us compare the contents of the following files
imports_olsrr.txtimports_blorr.txt
The files contain the names of R packages imported by the olsrr and blorr packages respectively (Full disclosure: both the above R pakages are developed by Rsquared Academy.).
diff uses certain special symbols and gives instructions to make the files identical. The instructions are on how to change the first file to make it identical to the second file. We list the symbols below
- a for add
- c for change
- d for delete
We will use the -w option to ignore white spaces while comparing the files.
diff -w imports_olsrr.txt imports_blorr.txt## 1a2
## > caret
## 3d3
## < cli
## 4a5
## > cliLet us interpret the results. 4a5 indicates after line 4 in file 1, add line 5 from file 2 to make both the files identical i.e. add caret which is line 5 in imports_blorr.txt after line 4 in imports_olsrr.txt which will make both the files identical.
Let us change the file order and see the instructions from diff.
diff -w imports_blorr.txt imports_olsrr.txt## 2d1
## < caret
## 4d2
## < clisymbols
## 5a4
## > clisymbols5d4 indicates delete line 5 from file 1 to match both the files at line4 i.e. delete caret which is line 5 in imports_blorr.txt to make both the files identical.
Side By Side
To view the differences between the files side by side, use the -y option.
diff -y imports_olsrr.txt imports_blorr.txt## car
| car
## checkmate
| caret
## cli
| checkmate
## clisymbols
| clisymbols
## > cliUnified Format
To view the differences between the files in a unified format, use the -u option.
diff -u imports_olsrr.txt imports_blorr.txt## --- imports_olsrr.txt 2019-09-20 13:36:03.000000000 +0530
## +++ imports_blorr.txt 2019-09-20 13:36:35.000000000 +0530
## @@ -1,4 +1,5 @@
## -car
## -checkmate
## -cli
## -clisymbols
## +car
## +caret
## +checkmate
## +clisymbols
## +cliCompare Recursively
To compare recursively, use the -r option. Let us compare the mypackage and myproject folders.
diff -r mypackage myproject## Only in mypackage: .Rbuildignore
## Only in mypackage: DESCRIPTION
## Only in mypackage: LICENSE
## Only in mypackage: NAMESPACE
## Only in mypackage: NEWS.md
## Only in mypackage: R
## Only in myproject/data: processed
## Only in myproject/data: raw
## Only in mypackage: docs
## Only in mypackage: man
## Only in myproject: output
## Only in myproject: run_analysis.R
## Only in mypackage: tests
## Only in mypackage: vignettesFile Details
To compare directories and view the names of files that differ, use the -rq option. In the below example, we look at the names of files that differ in mypackage and myproject folders.
diff -rq mypackage myproject## Only in mypackage: .Rbuildignore
## Only in mypackage: DESCRIPTION
## Only in mypackage: LICENSE
## Only in mypackage: NAMESPACE
## Only in mypackage: NEWS.md
## Only in mypackage: R
## Only in myproject/data: processed
## Only in myproject/data: raw
## Only in mypackage: docs
## Only in mypackage: man
## Only in myproject: output
## Only in myproject: run_analysis.R
## Only in mypackage: tests
## Only in mypackage: vignettesR Functions
In R, file operations can be performed using functions from both base R and the fs package.
| Command | R |
|---|---|
touch
|
file.create() / fs::file_create() / fs::file_touch()
|
cp
|
file.copy() / fs::file_copy() / fs::dir_copy()
|
mv
|
file.rename() / fs::file_move()
|
rm
|
file.remove() / fs::file_delete()
|
diff
|
Input/Output
In this section, we will explore commands that will
- display messages
- print file contents
- sort file contents
| Command | Description |
|---|---|
echo
|
Display messages |
cat
|
Print contents of a file |
head
|
Prints first ten lines of a file by default |
tail
|
Prints last ten lines of a file by default |
more
|
Open a file for interactive reading, scrolling & searching |
less
|
Open a file for interactive reading, scrolling & searching |
sort
|
Sort a file in ascending order |
Display Messages
The echo command prints text to the terminal. It can be used for writing or appending messages to a file as well.
| Command | Description |
|---|---|
echo
|
Display messages |
echo -n
|
Print message without trailing new line |
echo > file
|
Write message to a file |
echo >> file
|
Append message to a file |
echo -e
|
Enable interpretation of special characters |
Print Message
Let us start with a simple example. We will print the text Action of the Toes to the terminal. It is the release name for R version 3.6.1.
echo Action of the ToesRedirect Output
What if we want to redirect the output? Instead of printing the text to the terminal, we want to write it to a file. In such cases, use > along with the file name to redirect the output to the file. Keep in mind that > will overwrite files. If you want to append to files instead of overwriting, use >>.
echo Great Truth > release.txtPrint & Concatenate Files
The cat command reads data from files, and outputs their contents. It is the simplest way to display the contents of a file at the command line. It can be used to overwrite or append new content to files as well. cat stands for catenate and can be used to
- display text files
- copy text files into a new document
- append the contents of a text file to the end of another text file, combining them
| Command | Description |
|---|---|
cat
|
Print & concatenate files |
cat >
|
Concatenate several files into the target file |
cat >>
|
Append several files into the target file |
cat -n
|
Number all output lines |
Print Content
Let us print the content of the release_names.txt file (it contains R release names).
cat release_names.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeNumber All Output Lines
If you want to number the output line, use the -n option.
cat -n release_names.txt## 1 Unsuffered Consequences
## 2 Great Pumpkin
## 3 December Snowflakes
## 4 Gift-Getting Season
## 5 Easter Beagle
## 6 Roasted Marshmallows
## 7 Trick or Treat
## 8 Security Blanket
## 9 Masked Marvel
## 10 Good Sport
## 11 Frisbee Sailing
## 12 Warm Puppy
## 13 Spring Dance
## 14 Sock it to Me
## 15 Pumpkin Helmet
## 16 Smooth Sidewalk
## 17 Full of Ingredients
## 18 World-Famous Astronaut
## 19 Fire Safety
## 20 Wooden Christmas Tree
## 21 Very Secure Dishes
## 22 Very, Very Secure Dishes
## 23 Supposedly Educational
## 24 Bug in Your Hair
## 25 Sincere Pumpkin Patch
## 26 Another Canoe
## 27 You Stupid Darkness
## 28 Single Candle
## 29 Short Summer
## 30 Kite Eating TreeConcatenate Several Files
To concatenate the contents of several files into a target file, use >. In the below example, we concatenate the contents of the files release_names_18.txt and release_names_19.txt into a single file release_names_18_19.txt. In this case we are not printing the contents of the file to the terminal and instead we concatenate the contents from both the files and redirect the output to the target file.
cat release_names_18.txt release_names_19.txt > release_names_18_19.txt
cat release_names_18_19.txt## Someone to Lean On
## Joy in Playing
## Feather Spray
## Eggshell IglooGreat Truth
## Planting of a Tree
## Action of the ToesHead
The head command will display the firt 10 lines of a file(s) by default. It can be used to display the first few lines or bytes of a file as well.
| Command | Description |
|---|---|
head
|
Output the first parts of a file |
head -n
|
Output the first n lines of a file |
head -c
|
Output the first c bytes of a file |
head -n -x
|
Output everything but the last x lines of a file |
head -c -x
|
Output everything but the last x bytes of a file |
Output the first parts of a file
Let us use head to display the first 10 lines of the release_names.txt file.
head release_names.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good SportOutput the first n lines of a file
Using the n option, we can specify the number of lines to be displayed. In the below example, we display the first 5 lines.
head -n 5 release_names.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter BeagleOutput the first c bytes of a file
The c option can be used to display characters or bytes instead of lines. Let us display the first 5 bytes of the release_names.txt file.
head -c 5 release_names.txt## UnsufOutput everything but the last 5 lines of a file
To display the last parts of a file, use - while specifying the number of lines. In the below example, we display the last 5 lines of the file.
head -n -5 release_names.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin PatchOutput everything but the last 3 bytes of a file
In this example, we display the last 3 bytes of the file using the c option and - while specifying the number of bytes.
head -c -3 release_names.txt## Unsuffered Consequences
## Great Pumpkin
## December Snowflakes
## Gift-Getting Season
## Easter Beagle
## Roasted Marshmallows
## Trick or Treat
## Security Blanket
## Masked Marvel
## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreTail
The tail command displays the last 10 lines of a file(s) by default. It can be used to display the last few lines or bytes of a file as well.
| Command | Description |
|---|---|
tail
|
Display the last part of a file |
tail -n num
|
Show the last num lines of a file |
tail -n +num
|
Show all contents of the file starting from num line |
tail -c num
|
Show last num bytes of a file |
tail -f
|
Keep reading file until Ctrl + C |
tail -F
|
Keep reading file until Ctrl + C; even if the file is rotated |
Display the last parts of a file
Let us use tail to display the last 10 lines of the file.
tail release_names.txt## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeDisplay the last 5 lines of a file
As we did in the previous section, use n to specify the number of lines to be displayed.
tail -n 5 release_names.txt## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeDisplay all contents from line 10
We can use tail to display all contents of a file starting from a specific line. In the below example, we display all contents of the file starting from the 10th line using the n option and + prefix while specifying the number of lines.
tail -n +10 release_names.txt## Good Sport
## Frisbee Sailing
## Warm Puppy
## Spring Dance
## Sock it to Me
## Pumpkin Helmet
## Smooth Sidewalk
## Full of Ingredients
## World-Famous Astronaut
## Fire Safety
## Wooden Christmas Tree
## Very Secure Dishes
## Very, Very Secure Dishes
## Supposedly Educational
## Bug in Your Hair
## Sincere Pumpkin Patch
## Another Canoe
## You Stupid Darkness
## Single Candle
## Short Summer
## Kite Eating TreeDisplay the last 10 bytes of a file
Use the c option to display the last 7 bytes of a file.
tail -c 7 release_names.txt## TreeMore
The more command displays text, one screen at a time. It opens a file for
- interactive reading
- scrolling
- and searching
Press space to scroll down the page, the forward slash (/) for searching strings, n to go to the next match and q to quit.
| Command | Description |
|---|---|
more
|
Open a file for interactive reading, scrolling & searching |
space
|
Page down |
/
|
Search for a string; press n to go the next match
|
q
|
Quit |
Less
The less command is similar to more but offers more features. It allows the
user to scroll up and down, go to the beggining and end of the file, forward and backward search and the ability to go the next and previous match while searching the file.
| Command | Description |
|---|---|
less
|
Open a file for interactive reading, scrolling & searching |
space
|
Page down |
b
|
Page up |
G
|
Go to the end of file |
g
|
Go to the start of file |
/
|
Forward search |
?
|
Backward search |
n
|
Go to next match |
N
|
Go to previous match |
q
|
Quit |
Sort
The sort command will sort the contents of text file, line by line. Using additional options, we can
- sort a file in ascending/descending order
- ignore case while sorting
- use numeric order for sorting
- preserve only unique lines while sorting
Using the sort command, the contents can be sorted numerically and alphabetically. By default, the rules for sorting are:
- lines starting with a number will appear before lines starting with a letter.
- lines starting with a letter that appears earlier in the alphabet will appear before lines starting with a letter that appears later in the alphabet.
- lines starting with a lowercase letter will appear before lines starting with the same letter in uppercase.
Using additional options, the rules for sorting can be changed. We list the options in the below table.
| Command | Description |
|---|---|
sort
|
Sort lines of text files |
sort -r
|
Sort a file in descending order |
sort --ignore-case
|
Ignore case while sorting |
sort -n
|
Use numeric order for sorting |
sort -u
|
Preserve only unique lines while sorting |
Sort
Let us sort the contents of the pkg_names.txt file. It contains names R packages randomly selected from CRAN.
sort pkg_names.txt## ASIP
## AdMit
## AnalyzeTS
## AzureStor
## AzureStor
## BIGDAWG
## BIOMASS
## BIOMASS
## BenfordTests
## BinOrdNonNor
## BioCircos
## ClimMobTools
## CombinePValue
## Eagle
## FField
## ICAOD
## MARSS
## MIAmaxent
## MIAmaxent
## MIAmaxent
## MVB
## MVTests
## MaXact
## MaxentVariableSelection
## OptimaRegion
## OxyBS
## PathSelectMP
## PropScrRand
## RJDBC
## RPyGeo
## SCRT
## SMARTp
## SPEDInstabR
## SemiParSampleSel
## SetMethods
## SmallCountRounding
## SpatioTemporal
## SphericalK
## SuppDists
## Survgini
## TIMP
## TSeriesMMA
## VineCopula
## WGScan
## WPKDE
## accept
## accept
## addhaz
## alfr
## aweek
## aweek
## bayesbio
## blink
## breakfast
## cbsem
## corclass
## crsra
## cyclocomp
## dagitty
## disparityfilter
## edfReader
## errorlocate
## expstudies
## fermicatsR
## foretell
## gLRTH
## gazepath
## generalhoslem
## geoknife
## hdnom
## hindexcalculator
## ibd
## interplot
## kfigr
## logNormReg
## ltxsparklines
## lue
## mbir
## mcmcabn
## mev
## mgcViz
## mined
## mlflow
## mongolite
## mongolite
## mvShapiroTest
## odk
## overlapping
## pAnalysis
## pls
## pmdplyr
## poisbinom
## randtests
## redcapAPI
## rgw
## rless
## rsed
## rstudioapi
## solitude
## splithalfr
## sspline
## sybilccFBA
## tailr
## tailr
## tictactoe
## viridisLite
## vqtl
## widyr
## widyrDescending Order
Using the -r option which stands for --reverse the contents of the file can be sorted in descending/reverse order. Let us now sort the contents of the pkg_names.txt file in reverse order.
sort -r pkg_names.txt## widyr
## widyr
## vqtl
## viridisLite
## tictactoe
## tailr
## tailr
## sybilccFBA
## sspline
## splithalfr
## solitude
## rstudioapi
## rsed
## rless
## rgw
## redcapAPI
## randtests
## poisbinom
## pmdplyr
## pls
## pAnalysis
## overlapping
## odk
## mvShapiroTest
## mongolite
## mongolite
## mlflow
## mined
## mgcViz
## mev
## mcmcabn
## mbir
## lue
## ltxsparklines
## logNormReg
## kfigr
## interplot
## ibd
## hindexcalculator
## hdnom
## geoknife
## generalhoslem
## gazepath
## gLRTH
## foretell
## fermicatsR
## expstudies
## errorlocate
## edfReader
## disparityfilter
## dagitty
## cyclocomp
## crsra
## corclass
## cbsem
## breakfast
## blink
## bayesbio
## aweek
## aweek
## alfr
## addhaz
## accept
## accept
## WPKDE
## WGScan
## VineCopula
## TSeriesMMA
## TIMP
## Survgini
## SuppDists
## SphericalK
## SpatioTemporal
## SmallCountRounding
## SetMethods
## SemiParSampleSel
## SPEDInstabR
## SMARTp
## SCRT
## RPyGeo
## RJDBC
## PropScrRand
## PathSelectMP
## OxyBS
## OptimaRegion
## MaxentVariableSelection
## MaXact
## MVTests
## MVB
## MIAmaxent
## MIAmaxent
## MIAmaxent
## MARSS
## ICAOD
## FField
## Eagle
## CombinePValue
## ClimMobTools
## BioCircos
## BinOrdNonNor
## BenfordTests
## BIOMASS
## BIOMASS
## BIGDAWG
## AzureStor
## AzureStor
## AnalyzeTS
## AdMit
## ASIPIgnore case
To ignore case while sorting contents, use the --ignore-case option. Time to sort the pkg_names.txt file while ignoring case.
sort --ignore-case pkg_names.txt## accept
## accept
## addhaz
## AdMit
## alfr
## AnalyzeTS
## ASIP
## aweek
## aweek
## AzureStor
## AzureStor
## bayesbio
## BenfordTests
## BIGDAWG
## BinOrdNonNor
## BioCircos
## BIOMASS
## BIOMASS
## blink
## breakfast
## cbsem
## ClimMobTools
## CombinePValue
## corclass
## crsra
## cyclocomp
## dagitty
## disparityfilter
## Eagle
## edfReader
## errorlocate
## expstudies
## fermicatsR
## FField
## foretell
## gazepath
## generalhoslem
## geoknife
## gLRTH
## hdnom
## hindexcalculator
## ibd
## ICAOD
## interplot
## kfigr
## logNormReg
## ltxsparklines
## lue
## MARSS
## MaXact
## MaxentVariableSelection
## mbir
## mcmcabn
## mev
## mgcViz
## MIAmaxent
## MIAmaxent
## MIAmaxent
## mined
## mlflow
## mongolite
## mongolite
## MVB
## mvShapiroTest
## MVTests
## odk
## OptimaRegion
## overlapping
## OxyBS
## pAnalysis
## PathSelectMP
## pls
## pmdplyr
## poisbinom
## PropScrRand
## randtests
## redcapAPI
## rgw
## RJDBC
## rless
## RPyGeo
## rsed
## rstudioapi
## SCRT
## SemiParSampleSel
## SetMethods
## SmallCountRounding
## SMARTp
## solitude
## SpatioTemporal
## SPEDInstabR
## SphericalK
## splithalfr
## sspline
## SuppDists
## Survgini
## sybilccFBA
## tailr
## tailr
## tictactoe
## TIMP
## TSeriesMMA
## VineCopula
## viridisLite
## vqtl
## WGScan
## widyr
## widyr
## WPKDENumeric Order
To sort numerically, use the -n option which stands for --numeric-sort. In this example, we will use a different file, package_names.txt where the package names are prefixed by random numbers between 1 and 100.
sort -n package_names.txt## 1. cyclocomp
## 2. odk
## 3. redcapAPI
## 4. TIMP
## 5. pls
## 6. BinOrdNonNor
## 7. bayesbio
## 8. MVTests
## 9. pAnalysis
## 10. aweek
## 11. hdnom
## 12. ltxsparklines
## 13. MaXact
## 14. RJDBC
## 15. MIAmaxent
## 16. randtests
## 17. ASIP
## 18. gazepath
## 19. mcmcabn
## 20. rless
## 21. corclass
## 22. vqtl
## 23. disparityfilter
## 24. SCRT
## 25. RPyGeo
## 26. blink
## 27. gLRTH
## 28. splithalfr
## 29. sspline
## 29. sspline
## 30. logNormReg
## 31. BIGDAWG
## 31. BIGDAWG
## 32. SPEDInstabR
## 33. tailr
## 33. tailr
## 34. ibd
## 35. fermicatsR
## 36. mlflow
## 37. CombinePValue
## 38. BenfordTests
## 39. mev
## 40. MaxentVariableSelection
## 41. rstudioapi
## 42. OptimaRegion
## 43. accept
## 44. expstudies
## 45. solitude
## 45. solitude
## 46. cbsem
## 47. SMARTp
## 48. geoknife
## 49. SemiParSampleSel
## 50. mbir
## 51. interplot
## 52. ClimMobTools
## 53. MVB
## 54. OxyBS
## 55. hindexcalculator
## 56. MARSS
## 57. generalhoslem
## 58. alfr
## 59. AdMit
## 60. Eagle
## 61. PropScrRand
## 62. lue
## 63. dagitty
## 64. viridisLite
## 65. mined
## 65. mined
## 66. SuppDists
## 67. tictactoe
## 68. AzureStor
## 68. AzureStor
## 69. FField
## 70. rsed
## 70. rsed
## 71. kfigr
## 72. overlapping
## 72. overlapping
## 73. VineCopula
## 74. crsra
## 75. pmdplyr
## 76. errorlocate
## 77. SetMethods
## 78. sybilccFBA
## 79. mvShapiroTest
## 80. SpatioTemporal
## 81. mgcViz
## 82. breakfast
## 83. WPKDE
## 84. BIOMASS
## 85. edfReader
## 86. mongolite
## 87. WGScan
## 88. SphericalK
## 89. foretell
## 90. widyr
## 91. rgw
## 92. BioCircos
## 93. PathSelectMP
## 94. ICAOD
## 95. TSeriesMMA
## 96. poisbinom
## 97. AnalyzeTS
## 98. SmallCountRounding
## 99. Survgini
## 100. addhazPreserve Only Unique Lines
The -u option which stands for --unique will preserve only unique lines while sorting the contents of the file. In the below example, we remove all duplicate lines from the pkg_names.txt while sorting.
sort -u pkg_names.txt## ASIP
## AdMit
## AnalyzeTS
## AzureStor
## BIGDAWG
## BIOMASS
## BenfordTests
## BinOrdNonNor
## BioCircos
## ClimMobTools
## CombinePValue
## Eagle
## FField
## ICAOD
## MARSS
## MIAmaxent
## MVB
## MVTests
## MaXact
## MaxentVariableSelection
## OptimaRegion
## OxyBS
## PathSelectMP
## PropScrRand
## RJDBC
## RPyGeo
## SCRT
## SMARTp
## SPEDInstabR
## SemiParSampleSel
## SetMethods
## SmallCountRounding
## SpatioTemporal
## SphericalK
## SuppDists
## Survgini
## TIMP
## TSeriesMMA
## VineCopula
## WGScan
## WPKDE
## accept
## accept
## addhaz
## alfr
## aweek
## bayesbio
## blink
## breakfast
## cbsem
## corclass
## crsra
## cyclocomp
## dagitty
## disparityfilter
## edfReader
## errorlocate
## expstudies
## fermicatsR
## foretell
## gLRTH
## gazepath
## generalhoslem
## geoknife
## hdnom
## hindexcalculator
## ibd
## interplot
## kfigr
## logNormReg
## ltxsparklines
## lue
## mbir
## mcmcabn
## mev
## mgcViz
## mined
## mlflow
## mongolite
## mvShapiroTest
## odk
## overlapping
## pAnalysis
## pls
## pmdplyr
## poisbinom
## randtests
## redcapAPI
## rgw
## rless
## rsed
## rstudioapi
## solitude
## splithalfr
## sspline
## sybilccFBA
## tailr
## tictactoe
## viridisLite
## vqtl
## widyrWord Count
wc will print newline, word, and byte counts for file(s). If more than one file is specified, it will also print total line.
Count words, bytes and lines
wc release_names.txt## 30 73 546 release_names.txtCount lines in a file
wc -l release_names.txt## 30 release_names.txtCount words in a file
wc -w release_names.txt## 73 release_names.txtSearch & Regular Expressions
In this section, we will explore commands that will
- search for a given string in a file
- find files using names
- and search for binary executable files
| Command | Description |
|---|---|
grep
|
Search for a given string in a file |
find
|
Find files using filenames |
which
|
Search for binary executable files |
grep
The grep command is used for pattern matching. Along with additional options, it can be used to
- match pattern in input text
- ignore case
- search recursively for an exact string
- print filename and line number for each match
- invert match for excluding specific strings
grep processes text line by line, and prints any lines which match a specified pattern. grep, which stands for global regular expression print is a powerful tool for matching a regular expression against text in a file, multiple files, or a stream of input.
| Command | Description |
|---|---|
grep
|
Matches pattern in input text |
grep -i
|
Ignore case |
grep -RI
|
Search recursively for an exact string |
grep -E
|
Use extended regular expression |
grep -Hn
|
Print file name & corresponding line number for each match |
grep -v
|
Invert match for excluding specific strings |
Match Pattern in Input Text
Using grep let us search for packages that inlcude the letter R in their names.
grep R package_names.txt## 14. RJDBC
## 30. logNormReg
## 27. gLRTH
## 35. fermicatsR
## 42. OptimaRegion
## 61. PropScrRand
## 25. RPyGeo
## 47. SMARTp
## 24. SCRT
## 56. MARSS
## 85. edfReader
## 32. SPEDInstabR
## 98. SmallCountRoundingIgnore Case
In the previous case, grep returned only those packages whose name included R but not r i.e. it did not ignore the case of the letter. Using the -i option, we will now search while ignoring the case of the letter.
grep -i R package_names.txt## 14. RJDBC
## 58. alfr
## 64. viridisLite
## 99. Survgini
## 30. logNormReg
## 27. gLRTH
## 71. kfigr
## 72. overlapping
## 90. widyr
## 33. tailr
## 40. MaxentVariableSelection
## 33. tailr
## 72. overlapping
## 16. randtests
## 12. ltxsparklines
## 91. rgw
## 35. fermicatsR
## 21. corclass
## 68. AzureStor
## 42. OptimaRegion
## 61. PropScrRand
## 74. crsra
## 80. SpatioTemporal
## 23. disparityfilter
## 49. SemiParSampleSel
## 76. errorlocate
## 88. SphericalK
## 28. splithalfr
## 89. foretell
## 25. RPyGeo
## 50. mbir
## 51. interplot
## 6. BinOrdNonNor
## 47. SMARTp
## 38. BenfordTests
## 79. mvShapiroTest
## 92. BioCircos
## 55. hindexcalculator
## 41. rstudioapi
## 57. generalhoslem
## 24. SCRT
## 95. TSeriesMMA
## 82. breakfast
## 56. MARSS
## 70. rsed
## 68. AzureStor
## 85. edfReader
## 20. rless
## 75. pmdplyr
## 32. SPEDInstabR
## 3. redcapAPI
## 70. rsed
## 98. SmallCountRoundingHighlight
The --color option will highlight the matched strings.
grep -i --color R package_names.txt## 14. RJDBC
## 58. alfr
## 64. viridisLite
## 99. Survgini
## 30. logNormReg
## 27. gLRTH
## 71. kfigr
## 72. overlapping
## 90. widyr
## 33. tailr
## 40. MaxentVariableSelection
## 33. tailr
## 72. overlapping
## 16. randtests
## 12. ltxsparklines
## 91. rgw
## 35. fermicatsR
## 21. corclass
## 68. AzureStor
## 42. OptimaRegion
## 61. PropScrRand
## 74. crsra
## 80. SpatioTemporal
## 23. disparityfilter
## 49. SemiParSampleSel
## 76. errorlocate
## 88. SphericalK
## 28. splithalfr
## 89. foretell
## 25. RPyGeo
## 50. mbir
## 51. interplot
## 6. BinOrdNonNor
## 47. SMARTp
## 38. BenfordTests
## 79. mvShapiroTest
## 92. BioCircos
## 55. hindexcalculator
## 41. rstudioapi
## 57. generalhoslem
## 24. SCRT
## 95. TSeriesMMA
## 82. breakfast
## 56. MARSS
## 70. rsed
## 68. AzureStor
## 85. edfReader
## 20. rless
## 75. pmdplyr
## 32. SPEDInstabR
## 3. redcapAPI
## 70. rsed
## 98. SmallCountRoundingPrint Filename
If there is more than one file to search, use the -H option to print the filename for each match.
grep -i --color -H bio package_names.txt## package_names.txt:84. BIOMASS
## package_names.txt:92. BioCircos
## package_names.txt:7. bayesbioPrint Corresponding Line Number
The -n option will print the corresponding line number of the match in the file.
grep -i --color -n bio package_names.txt## 59:84. BIOMASS
## 71:92. BioCircos
## 88:7. bayesbioPrint Filename & Line Number
Let us print both the file name and the line number for each match.
grep -i --color -Hn R package_names.txt## package_names.txt:1:14. RJDBC
## package_names.txt:3:58. alfr
## package_names.txt:8:64. viridisLite
## package_names.txt:14:99. Survgini
## package_names.txt:15:30. logNormReg
## package_names.txt:16:27. gLRTH
## package_names.txt:18:71. kfigr
## package_names.txt:20:72. overlapping
## package_names.txt:21:90. widyr
## package_names.txt:22:33. tailr
## package_names.txt:23:40. MaxentVariableSelection
## package_names.txt:26:33. tailr
## package_names.txt:27:72. overlapping
## package_names.txt:30:16. randtests
## package_names.txt:31:12. ltxsparklines
## package_names.txt:32:91. rgw
## package_names.txt:33:35. fermicatsR
## package_names.txt:37:21. corclass
## package_names.txt:38:68. AzureStor
## package_names.txt:41:42. OptimaRegion
## package_names.txt:42:61. PropScrRand
## package_names.txt:43:74. crsra
## package_names.txt:51:80. SpatioTemporal
## package_names.txt:52:23. disparityfilter
## package_names.txt:54:49. SemiParSampleSel
## package_names.txt:55:76. errorlocate
## package_names.txt:57:88. SphericalK
## package_names.txt:61:28. splithalfr
## package_names.txt:62:89. foretell
## package_names.txt:63:25. RPyGeo
## package_names.txt:64:50. mbir
## package_names.txt:65:51. interplot
## package_names.txt:66:6. BinOrdNonNor
## package_names.txt:67:47. SMARTp
## package_names.txt:68:38. BenfordTests
## package_names.txt:69:79. mvShapiroTest
## package_names.txt:71:92. BioCircos
## package_names.txt:75:55. hindexcalculator
## package_names.txt:78:41. rstudioapi
## package_names.txt:80:57. generalhoslem
## package_names.txt:84:24. SCRT
## package_names.txt:85:95. TSeriesMMA
## package_names.txt:87:82. breakfast
## package_names.txt:96:56. MARSS
## package_names.txt:97:70. rsed
## package_names.txt:98:68. AzureStor
## package_names.txt:100:85. edfReader
## package_names.txt:101:20. rless
## package_names.txt:102:75. pmdplyr
## package_names.txt:103:32. SPEDInstabR
## package_names.txt:104:3. redcapAPI
## package_names.txt:106:70. rsed
## package_names.txt:107:98. SmallCountRoundingInvert Match
Use the -v option to select non-matching lines. In the below example, we search for packages whose name does not include R while ignoring the case.
grep -v -i R package_names.txt## 36. mlflow
## 10. aweek
## 31. BIGDAWG
## 22. vqtl
## 29. sspline
## 39. mev
## 66. SuppDists
## 15. MIAmaxent
## 31. BIGDAWG
## 29. sspline
## 60. Eagle
## 83. WPKDE
## 11. hdnom
## 26. blink
## 18. gazepath
## 52. ClimMobTools
## 44. expstudies
## 65. mined
## 81. mgcViz
## 45. solitude
## 9. pAnalysis
## 65. mined
## 94. ICAOD
## 48. geoknife
## 45. solitude
## 67. tictactoe
## 46. cbsem
## 93. PathSelectMP
## 96. poisbinom
## 17. ASIP
## 5. pls
## 84. BIOMASS
## 59. AdMit
## 77. SetMethods
## 53. MVB
## 2. odk
## 86. mongolite
## 4. TIMP
## 97. AnalyzeTS
## 87. WGScan
## 63. dagitty
## 69. FField
## 13. MaXact
## 73. VineCopula
## 7. bayesbio
## 34. ibd
## 8. MVTests
## 19. mcmcabn
## 43. accept
## 78. sybilccFBA
## 62. lue
## 100. addhaz
## 37. CombinePValue
## 1. cyclocomp
## 54. OxyBSRecursive Search
Use the -r option to search recursively. In the below example, we search all files with the .txt extension for the string bio while ignoring the case.
grep -i --color -r bio *.txt## package_names.txt:84. BIOMASS
## package_names.txt:92. BioCircos
## package_names.txt:7. bayesbio
## pkg_names.txt:BIOMASS
## pkg_names.txt:BioCircos
## pkg_names.txt:BIOMASS
## pkg_names.txt:bayesbiofind
The find command can be used for searching files and directories. Using additional options, we can
- search files by extension type
- ignore case while searching files/directories
find is a powerful tool for working with the files. It can be used on its own to locate files, or in conjunction with other programs to perform operations on those files.
| Command | Description |
|---|---|
find
|
Find files or directories under the given directory; recursively |
find -name '*.txt'
|
Find files by extension |
find -type d -iname
|
Find directories matching a given name, in case-insensitive mode |
find -type d -name
|
Find directories matching a given name, in case-sensitive mode |
Search Recursively
Let us use find to search for the file release_names.txt recursively. The -name option is used to specify the name of the file we are searching.
find -name release_names.txt## ./release_names.txt
## ./r_releases/release_names.txtThere are two files with the name release_names.txt present in the current working directory and in r_releases directory.
Search by Extension
Let us search for all files with .txt extension in the r_releases folder.
find r_releases -name '*.txt'## r_releases/release_names.txt
## r_releases/release_names_2.txt
## r_releases/release_names_3.txtThere are 3 files with the .txt extension in r_releases folder.
Case-insensitive Mode
Search for all folders with the name R or r. Here we use the -iname option to ignore case while searching. The -type option is used to specify whether we are searching for files or folders. Since we are searching for folder/directory, we use it along with d i.e. directory to indicate that we are searching for directories and not files.
find -type d -iname R## ./mypackage/R
## ./rCase-sensitive Mode
Search for all folders with the name r. It should exclude any folder with the name R.
find -type d -name r## ./rData Transfer & Network
In this section, we will explore commands that will allow us to download files from the internet.
| Command | Description |
|---|---|
wget
|
Download files from the web |
curl
|
Transfer data from or to a server |
hostname
|
Name of the current host |
ping
|
Ping a remote host |
nslookup
|
Name server details |
We have not executed the commands in this ebook as downloading multiple files from the internet will take a lot of time or result in errors but we have checked all the commands offline to ensure that they work.
wget
The wget command will download contents of a URL and files from the internet.
Using additional options, we can
- download contents/files to a file
- continue incomplete downloads
- download multiple files
- limit download speed and number of retries
| Command | Description |
|---|---|
wget url
|
Download contents of a url |
wget -o file url
|
Download contents of url to a file |
wget -c
|
Continue an incomplete download |
wget -P folder_name -i urls.txt
|
Download all urls stored in a text file to a specific directory |
wget --limit-rate
|
Limit download speed |
wget --tries
|
Limit number of retries |
wget --quiet
|
Turn off output |
wget --no-verbose
|
Print basic information |
wget --progress-dot
|
Change progress bar type to dot |
wget --timestamping
|
Check if the timestamp of the file has changed before downloading |
wget --wait
|
Wait between retrievals |
Download URL
Let us first use wget to download contents of a URL. Note, we are not downloading file as such but just the content of the URL. We will use the URL of the home page of R project.
wget https://www.r-project.org/If you look at the list of files, you can see a new file, index.html which we just downloaded using wget. Downloading contents this way will lead to confusion if we are dealing with multiple URLs. Let us learn to save the contents to a file (we can specify the name of the file which should help avoid confusion.)
Specify Filename
In this example, we download contents from the same URL and in addition specify the name of the file in which the content must be saved. Here we save it in a new file, rhomepage.html using the -o option followed by the filename.
wget -o rhomepage.html https://www.r-project.org/Download File
How about downloading a file instead of a URL? In this example, we will download a logfile from the RStudio CRAN mirror. It contains the details of R downloads and individual package downloads. If you are a package developer and would want to know the countries in which your packages are downloaded, you will find this useful. We will download the file for 29th September and save it as sep_29.csv.gz.
wget -o sep_29.csv.gz http://cran-logs.rstudio.com/2019/2019-09-29.csv.gzDownload Multiple URLs
How do we download multiple URLs? One way is to specify the URLs one after the other separated by a space or save all URLs in a file and read them one by one. In the below example, we have saved multiple URLs in the file urls.txt.
cat urls.txt## http://cran-logs.rstudio.com/2019/2019-09-26.csv.gz
## http://cran-logs.rstudio.com/2019/2019-09-27.csv.gz
## http://cran-logs.rstudio.com/2019/2019-09-28.csv.gzWe will download all the above URLs and save them in a new folder downloads. The -i indicates that the URLs must be read from a file (local or external). The -P option allows us to specify the directory into which all the files will be downloaded.
wget -P downloads -i urls.txt Quiet
The --quiet option will turn off wget output. It will not show any of the following details:
- name of the file being saved
- file size
- download speed
- eta etc.
wget –-quiet http://cran-logs.rstudio.com/2019/2019-10-06.csv.gzNo Verbose
Using the -nv or --no-verbose option, we can turn off verbose without being completely quiet (as we did in the previous example). Any error messages and basic information will still be printed.
wget –-no-verbose http://cran-logs.rstudio.com/2019/2019-10-13.csv.gz Check Timestamp
Let us say we have already downloaded a file from a URL. The file is updated from time to time and we intend to keep the local copy updated as well. Using the --timestamping option, the local file will have timestamp matching the remote file; if the remote file is not newer (not updated), no download will occur i.e. if the timestamp of the remote file has not changed it will not be downloaded. This is very useful in case of large files where you do not want to download them unless they have been updated.
wget –-timestamping http://cran-logs.rstudio.com/2019/2019-10-13.csv.gzcurl
The curl command will transfer data from or to a server. We will only look at
downloading files from the internet.
| Command | Description |
|---|---|
curl url
|
Download contents of a url |
curl url -o file
|
Download contents of url to a file |
curl url > file
|
Download contents of url to a file |
curl -s
|
Download in silent or quiet mode |
Download URL
Let us download the home page of the R project using curl.
curl https://www.r-project.org/Specify File
Let us download another log file from the RStudio CRAN mirror and save it into a file using the -o option.
curl http://cran-logs.rstudio.com/2019/2019-09-08.csv.gz -o sept_08.csv.gz Another way to save a downloaded file is to use > followed by the name of the file as shown in the below example.
curl http://cran-logs.rstudio.com/2019/2019-09-01.csv.gz > sep_01.csv.gzDownload Silently
The -s option will allow you to download files silently. It will mute curl and will not display progress meter or error messages.
curl http://cran-logs.rstudio.com/2019/2019-09-01.csv.gz -o sept_01.csv.gz -sR Functions
In R, we can use download.file() to download files from the internet. The following packages offer functionalities that you will find useful.
| Command | R |
|---|---|
wget
|
download.file()
|
curl
|
curl::curl_download()
|
hostname
|
R.utils::getHostname.System()
|
ping
|
pingr::ping()
|
nslookup
|
curl::nslookup()
|
sudo
sudo Super User DO is a prefix of commands that only superuser or root users are allowed to run. It is similar to run as administrator option in Windows. It is used to install, update and remove software. We will use it in the next section to install & update packages. If you are using RStudio Cloud, you will not be able to run sudo (users do not have root privileges).
| Command | Description |
|---|---|
dpkg --list
|
List installed packages |
sudo apt-get update
|
Update packages |
sudo apt-get install
|
Install packages |
sudo apt-get remove
|
Remove packages (retain configuration, plugins and settings |
sudo apt-get purge
|
Remove packages including personalized settings |
sudo apt-get autoremove
|
Remove any dependencies no longer in use |
File Compression
tar
The tar command is used for file compression. It works with both tar and
tar.gz extensions. It is used to
- list files
- extract files
- create archives
- append file to existing archives
tar creates, maintains, modifies, and extracts files that are archived in the tar format. Tar stands for tape archive and is an archiving file format.
| Command | Description |
|---|---|
tar tvf
|
List an archive |
tar tvfz
|
List a gzipped archive |
tar xvf
|
Extract an archive |
tar xvfz
|
Extract a gzipped archive |
tar cvf
|
Create an uncompressed tar archive |
tar cvfz
|
Create a tar gzipped archive |
tar rvf
|
Add a file to an existing archive |
tar rvfz
|
Add a file to an existing gzipped archive |
We will use different options along with the tar command for listing, extracting, creating and adding files. The vf (v stands for verbosely show .tar file progress and f stands for file name type of the archive file) option is common for all the above operations while the following are specific.
tfor listingxfor extractingcfor creatingrfor adding files
While dealing with tar.gz archives we will use z in addition to vf and the above options.
List
Let us list all the files & folders in release_names.tar. As mentioned above. to list the files in the archive, we use the t option.
tar -tvf release_names.tar ## -rwxrwxrwx aravind/aravind 546 2019-09-16 15:59 release_names.txt
## -rwxrwxrwx aravind/aravind 65 2019-09-16 15:58 release_names_18.txt
## -rwxrwxrwx aravind/aravind 53 2019-09-16 15:59 release_names_19.txtExtract
Let us extract files from release_names.tar using the x option in addition to vf.
tar -xvf release_names.tar
ls## release_names.txt
## release_names_18.txt
## release_names_19.txt
## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject3
## myproject4
## package_names.txt
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_18_19.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipAdd
To add a file to an existing archive, use the r option. Let us add release_names_18.txt and release_names_19.txt to the archive we created in the previous step.
tar -rvf release_names.tar release_names_18.txt release_names_19.txt## release_names_18.txt
## release_names_19.txtCreate
Using the c option we can create tar archives. In the below example, we are using a single file but you can specify multiple files and folders as well.
tar -cvf pkg_names.tar pkg_names.txt## pkg_names.txtgzip
| Command | Description |
|---|---|
gzip
|
Compress a file |
gzip -d
|
Decompress a file |
gzip -c
|
Compress a file and specify the output file name |
zip -r
|
Compress a directory |
zip
|
Add files to an existing zip file |
unzip
|
Extract files from a zip files |
unzip -d
|
Extract files from a zip file and specify the output file name |
unzip -l
|
List contents of a zip file |
gzip, gunzip, and zcat commands are used to compress or expand files in the GNU GZIP format i.e. files with .gz extension
Compress
Let us compress release_names.txt file using gzip.
gzip release_names.txt
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject3
## myproject4
## package_names.txt
## pkg_names.tar
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt.gz
## release_names_18.txt
## release_names_18_19.txt
## release_names_19.txt
## sept_15.csv.gz
## urls.txt
## zip_example.zipDecompress
Use the -d option with gzip to decompress a file. In the below example, we decompress the sept_15.csv.gz file (downloaded using wget or curl earlier). You can also use gunzip for the same result.
gzip -d sept_15.csv.gz
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject3
## myproject4
## package_names.txt
## pkg_names.tar
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_18_19.txt
## release_names_19.txt
## sept_15.csv
## urls.txt
## zip_example.zipSpecify Filename
Use -c and > to specify a different file name while compressing using gzip. In the below example, gzip will create releases.txt.gz instead of release_names.txt.gz.
gzip -c release_names.txt > releases.txt.gz
ls## analysis.R
## bash.R
## bash.sh
## imports_blorr.txt
## imports_olsrr.txt
## lorem-ipsum.txt
## main_project.zip
## myfiles
## mypackage
## myproject
## myproject3
## myproject4
## package_names.txt
## pkg_names.tar
## pkg_names.txt
## r
## r2
## r_releases
## release_names.tar
## release_names.tar.gz
## release_names.txt
## release_names_18.txt
## release_names_18_19.txt
## release_names_19.txt
## releases.txt.gz
## sept_15.csv
## urls.txt
## zip_example.zipzip & unzip
zip creates ZIP archives while unzip lists and extracts compressed files in a ZIP archive.
List
Let us list all the files and folders in main_project.zip() using unzip and the -l option.
unzip -l main_project.zip## Archive: main_project.zip
## Length Date Time Name
## --------- ---------- ----- ----
## 0 2019-09-23 18:07 myproject/
## 0 2019-09-20 14:02 myproject/.gitignore
## 0 2019-09-23 18:07 myproject/data/
## 0 2019-09-20 14:02 myproject/data/processed/
## 0 2019-09-20 14:02 myproject/data/raw/
## 0 2019-09-20 14:02 myproject/output/
## 0 2019-09-20 14:02 myproject/README.md
## 13 2019-09-20 14:02 myproject/run_analysis.R
## 0 2019-09-20 14:02 myproject/src/
## 0 2019-09-23 18:07 mypackage/
## 0 2019-09-20 14:11 mypackage/.gitignore
## 0 2019-09-20 14:11 mypackage/.Rbuildignore
## 0 2019-09-20 14:10 mypackage/data/
## 0 2019-09-20 14:11 mypackage/DESCRIPTION
## 0 2019-09-20 14:10 mypackage/docs/
## 0 2019-09-20 14:11 mypackage/LICENSE
## 0 2019-09-20 14:10 mypackage/man/
## 0 2019-09-20 14:11 mypackage/NAMESPACE
## 0 2019-09-20 14:11 mypackage/NEWS.md
## 0 2019-09-20 14:10 mypackage/R/
## 0 2019-09-20 14:11 mypackage/README.md
## 0 2019-09-20 14:11 mypackage/src/
## 0 2019-09-20 14:10 mypackage/tests/
## 0 2019-09-20 14:10 mypackage/vignettes/
## 0 2019-09-23 18:07 myfiles/
## 12 2019-09-20 15:30 myfiles/analysis.R
## 7 2019-09-20 15:31 myfiles/NEWS.md
## 9 2019-09-20 15:31 myfiles/README.md
## 546 2019-09-20 15:29 myfiles/release_names.txt
## 65 2019-09-20 15:29 myfiles/release_names_18.txt
## 53 2019-09-20 15:30 myfiles/release_names_19.txt
## 12 2019-09-20 15:30 myfiles/visualization.R
## 15333 2019-10-01 16:58 bash.sh
## 0 2019-09-16 12:42 r/
## --------- -------
## 16050 34 filesExtract
Using unzip, let us now extract files and folders from zip_example.zip.
unzip zip_example.zip## Archive: zip_example.zip
## creating: zip_example/
## inflating: zip_example/bash.sh
## inflating: zip_example/pkg_names.txtUsing the -d option, we can extract the contents of zip_example.zip to a specific folder. In the below example, we extract it to a new folder examples.
unzip zip_example.zip –d myexamples## [1] "Archive: zip_example.zip"
## [2] " creating: examples/zip_example/"
## [3] " inflating: examples/zip_example/bash.sh "
## [4] " inflating: examples/zip_example/pkg_names.txt "Compress
Use the -r option along with zip to create a ZIP archive. In the below example, we create a ZIP archive of myproject folder.
zip -r myproject.zip myproject
ls## adding: myproject/ (stored 0%)
## adding: myproject/.gitignore (stored 0%)
## adding: myproject/data/ (stored 0%)
## adding: myproject/data/processed/ (stored 0%)
## adding: myproject/data/raw/ (stored 0%)
## adding: myproject/output/ (stored 0%)
## adding: myproject/README.md (stored 0%)
## adding: myproject/run_analysis.R (stored 0%)
## adding: myproject/src/ (stored 0%)We can compress multiple directories using zip. The names of the directories must be separated by a space as shown in the below example where we compress myproject and mypackage into a single ZIP archive.
zip -r packproj.zip myproject mypackage
ls## adding: myproject/ (stored 0%)
## adding: myproject/.gitignore (stored 0%)
## adding: myproject/data/ (stored 0%)
## adding: myproject/data/processed/ (stored 0%)
## adding: myproject/data/raw/ (stored 0%)
## adding: myproject/output/ (stored 0%)
## adding: myproject/README.md (stored 0%)
## adding: myproject/run_analysis.R (stored 0%)
## adding: myproject/src/ (stored 0%)
## adding: mypackage/ (stored 0%)
## adding: mypackage/.gitignore (stored 0%)
## adding: mypackage/.Rbuildignore (stored 0%)
## adding: mypackage/data/ (stored 0%)
## adding: mypackage/DESCRIPTION (stored 0%)
## adding: mypackage/docs/ (stored 0%)
## adding: mypackage/LICENSE (stored 0%)
## adding: mypackage/man/ (stored 0%)
## adding: mypackage/NAMESPACE (stored 0%)
## adding: mypackage/NEWS.md (stored 0%)
## adding: mypackage/R/ (stored 0%)
## adding: mypackage/README.md (stored 0%)
## adding: mypackage/src/ (stored 0%)
## adding: mypackage/tests/ (stored 0%)
## adding: mypackage/vignettes/ (stored 0%)Add
To add a new file/folder to an existing archive, specify the name of the archive followed by the name of the file or the folder. In the below example, we add the bash.sh file to the myproject.zip archive created in a previous step.
zip myproject.zip bash.sh## adding: bash.sh (deflated 78%)R Functions
tar & tar.gz
In R, we can use the tar() and untar() functions from the utils package to handle .tar and .tar.gz archives.
| Command | R |
|---|---|
tar tvf
|
utils::untar('archive.tar', list = TRUE)
|
tar tvfz
|
utils::untar('archive.tar.gz', list = TRUE)
|
tar xvf
|
utils::untar('archive.tar')
|
tar xvfz
|
utils::untar('archive.tar.gz')
|
tar cvf
|
utils::tar('archive.tar')
|
tar cvfz
|
utils::tar('archive.tar', compression = 'gzip')
|
zip & gzip
The zip package has the functionalities to handle ZIP archives. The tar() and untar() functions from the utils package can handle GZIP archives.
| Command | R |
|---|---|
gzip
|
utils::tar(compression = 'gzip' / R.utils::gzip()
|
gzip -d
|
utils::untar() / R.utils::gunzip()
|
gzip -c
|
utils::untar(exdir = filename)
|
zip -r
|
zip::zip()
|
zip
|
zip::zipr_append()
|
unzip
|
zip::unzip()
|
unzip -d
|
zip::unzip(exdir = dir_name)
|
unzip -l
|
zip::zip_list()
|
System Info
In this section, we will explore commands that will allow us to
- display information about the system
- display memory usage information
- display file system disk space usage
- exit the terminal
- run commands a superuser
- shutdown the system
| Command | Description |
|---|---|
uname
|
Display important information about the system |
free
|
Display free, used, swap memory in the system |
df
|
Display file system disk space usage |
exit
|
Exit the terminal |
sudo
|
Run command as super user |
shutdown
|
Shutdown the system |
uname
The uname command is used to view important information about the system. Using additional options, we can
- print details about operating system
- hardware & software related information
| Command | Description |
|---|---|
uname
|
Print details about the current machine and the operating system running on it |
uname -mp
|
Hardware related information; machine & processor |
uname -srv
|
Software related information; operating system, release number and version |
uname -n
|
Nodename of the system |
uname -a
|
Print all available information system |
Print all available information about the system
uname -a## Linux Aravind 4.4.0-18362-Microsoft #836-Microsoft Mon May 05 16:04:00 PST 2020 x86_64 x86_64 x86_64 GNU/LinuxDisplay free, used, swap memory in the system
free## total used free shared buff/cache available
## Mem: 3621900 1815864 1576684 17720 229352 1672304
## Swap: 11010048 260640 10749408Display file system disk space usage
df## Filesystem 1K-blocks Used Available Use% Mounted on
## rootfs 188482144 142470276 46011868 76% /
## none 188482144 142470276 46011868 76% /dev
## none 188482144 142470276 46011868 76% /run
## none 188482144 142470276 46011868 76% /run/lock
## none 188482144 142470276 46011868 76% /run/shm
## none 188482144 142470276 46011868 76% /run/user
## cgroup 188482144 142470276 46011868 76% /sys/fs/cgroup
## C:\ 188482144 142470276 46011868 76% /mnt/c
## D:\ 18660348 17154308 1506040 92% /mnt/d
## F:\ 3196924 231756 2965168 8% /mnt/f
## G:\ 86383612 24561220 61822392 29% /mnt/g
## H:\ 86383612 14755988 71627624 18% /mnt/h
## J:\ 83185660 18797916 64387744 23% /mnt/jOthers
In this section, let us look at a few other useful commands that will allow us to
- see how long a command takes to execute
- delay activity
- display and clear command history list
| Command | Description |
|---|---|
time
|
See how long a command takes to execute |
sleep
|
Delay activity in seconds |
sleep 1m
|
Delay activity in minutes |
sleep 1h
|
Delay activity in hours |
history
|
Display command history list with line numbers |
history -c
|
Clear the command history list |
Funny Commands
Below are a few funny commands for you to try out. Use sudo apt-get install to install fortune and banner before trying them.
| Command | Description |
|---|---|
fortune
|
Poignant, inspirational & silly phrases |
yes
|
Output a string repeatedly until killed |
banner
|
ASCII banner |
rev
|
Reverse each character |
Execute Commands from R
Now, let us turn our attention to executing commands from R using system2().
Here we will focus on the following
- execute a command without arguments
- execute commands with arguments
- redirect output
Let us try to execute a command without any additional arguments. We will execute the ls command to list all files and directories. Use system2() and specify the command using the command argument. Whenever you are trying to execute a command from R, the first argument or input should be the command and it must be enclosed in quotes.
system2(command = "ls")## [1] "2017-02-05-variables.Rmd"
## [2] "2017-02-05-variables.html"
## [3] "2017-02-17-data-types-in-r.Rmd"
## [4] "2017-02-17-data-types-in-r.html"
## [5] "2017-03-01-getting-help-in-r.html"
## [6] "2017-03-13-beginners-guide-to-r-package-ecosystem.Rmd"
## [7] "2017-03-13-beginners-guide-to-r-package-ecosystem.html"
## [8] "2017-03-25-vectors.Rmd"
## [9] "2017-03-25-vectors.html"
## [10] "2017-03-29-vectors-part-2.Rmd"
## [11] "2017-03-29-vectors-part-2.html"
## [12] "2017-04-03-vectors-part-3.Rmd"
## [13] "2017-04-03-vectors-part-3.html"
## [14] "2017-04-06-matrix.Rmd"
## [15] "2017-04-06-matrix.html"
## [16] "2017-04-12-matrix-part-2.Rmd"
## [17] "2017-04-12-matrix-part-2.html"
## [18] "2017-04-18-lists.Rmd"
## [19] "2017-04-18-lists.html"
## [20] "2017-04-30-factors.Rmd"
## [21] "2017-04-30-factors.html"
## [22] "2017-05-12-dataframes.Rmd"
## [23] "2017-05-12-dataframes.html"
## [24] "2017-05-24-data-visualization-with-r-introduction.Rmd"
## [25] "2017-05-24-data-visualization-with-r-introduction.html"
## [26] "2017-06-05-data-visualization-with-r-title-and-axis-labels.Rmd"
## [27] "2017-06-05-data-visualization-with-r-title-and-axis-labels.html"
## [28] "2017-06-17-data-visualization-with-r-scatter-plots.Rmd"
## [29] "2017-06-17-data-visualization-with-r-scatter-plots.html"
## [30] "2017-06-29-data-visualization-with-r-line-graphs.Rmd"
## [31] "2017-06-29-data-visualization-with-r-line-graphs.html"
## [32] "2017-07-11-data-visualization-with-r-bar-plots.Rmd"
## [33] "2017-07-11-data-visualization-with-r-bar-plots.html"
## [34] "2017-07-23-data-visualization-with-r-box-plots.Rmd"
## [35] "2017-07-23-data-visualization-with-r-box-plots.html"
## [36] "2017-08-04-data-visualization-with-r-histogram.Rmd"
## [37] "2017-08-04-data-visualization-with-r-histogram.html"
## [38] "2017-08-16-data-visualization-with-r-legends.Rmd"
## [39] "2017-08-16-data-visualization-with-r-legends.html"
## [40] "2017-08-28-data-visualization-with-r-text-annotations.Rmd"
## [41] "2017-08-28-data-visualization-with-r-text-annotations.html"
## [42] "2017-09-09-data-visualization-with-r-combining-plots.Rmd"
## [43] "2017-09-09-data-visualization-with-r-combining-plots.html"
## [44] "2017-10-03-ggplot2-quick-tour.Rmd"
## [45] "2017-10-03-ggplot2-quick-tour.html"
## [46] "2017-10-15-ggplot2-introduction-to-geoms.Rmd"
## [47] "2017-10-15-ggplot2-introduction-to-geoms.html"
## [48] "2017-10-27-ggplot2-introduction-to-aesthetics.Rmd"
## [49] "2017-10-27-ggplot2-introduction-to-aesthetics.html"
## [50] "2017-11-08-ggplot2-axis-plot-labels.Rmd"
## [51] "2017-11-08-ggplot2-axis-plot-labels.html"
## [52] "2017-11-20-ggplot2-text-annotations.Rmd"
## [53] "2017-11-20-ggplot2-text-annotations.html"
## [54] "2017-12-02-ggplot2-scatter-plots.Rmd"
## [55] "2017-12-02-ggplot2-scatter-plots.html"
## [56] "2017-12-14-ggplot2-line-graphs.Rmd"
## [57] "2017-12-14-ggplot2-line-graphs.html"
## [58] "2017-12-26-ggplot2-bar-plots.Rmd"
## [59] "2017-12-26-ggplot2-bar-plots.html"
## [60] "2018-01-07-ggplot2-box-plots.Rmd"
## [61] "2018-01-07-ggplot2-box-plots.html"
## [62] "2018-01-19-ggplot2-histogram.Rmd"
## [63] "2018-01-19-ggplot2-histogram.html"
## [64] "2018-01-31-ggplot2-guides-axes.Rmd"
## [65] "2018-01-31-ggplot2-guides-axes.html"
## [66] "2018-02-12-ggplot2-guides-legends.Rmd"
## [67] "2018-02-12-ggplot2-guides-legends.html"
## [68] "2018-02-24-guides-legends-part-2.Rmd"
## [69] "2018-02-24-guides-legends-part-2.html"
## [70] "2018-03-08-legend-part-3.Rmd"
## [71] "2018-03-08-legend-part-3.html"
## [72] "2018-03-20-legend-part-4.Rmd"
## [73] "2018-03-20-legend-part-4.html"
## [74] "2018-04-01-legend-part-5.Rmd"
## [75] "2018-04-01-legend-part-5.html"
## [76] "2018-04-13-legend-part-6.Rmd"
## [77] "2018-04-13-legend-part-6.html"
## [78] "2018-04-25-ggplot2-facets-combine-multiple-plots.Rmd"
## [79] "2018-04-25-ggplot2-facets-combine-multiple-plots.html"
## [80] "2018-05-07-ggplot2-themes.Rmd"
## [81] "2018-05-07-ggplot2-themes.html"
## [82] "2018-07-30-importing-data-into-r-part-1.Rmd"
## [83] "2018-07-30-importing-data-into-r-part-1.html"
## [84] "2018-08-11-importing-data-into-r-part-2.Rmd"
## [85] "2018-08-11-importing-data-into-r-part-2.html"
## [86] "2018-08-23-data-wrangling-with-dplyr-part-1.Rmd"
## [87] "2018-08-23-data-wrangling-with-dplyr-part-1.html"
## [88] "2018-09-04-data-wrangling-with-dplyr-part-2.Rmd"
## [89] "2018-09-04-data-wrangling-with-dplyr-part-2.html"
## [90] "2018-09-16-data-wrangling-with-dplyr-part-3.Rmd"
## [91] "2018-09-16-data-wrangling-with-dplyr-part-3.html"
## [92] "2018-09-28-introduction-to-tibbles.Rmd"
## [93] "2018-09-28-introduction-to-tibbles.html"
## [94] "2018-10-10-readable-code-with-pipes.Rmd"
## [95] "2018-10-10-readable-code-with-pipes.html"
## [96] "2018-10-22-hacking-strings-with-stringr.Rmd"
## [97] "2018-10-22-hacking-strings-with-stringr.html"
## [98] "2018-11-03-working-with-dates-in-r.Rmd"
## [99] "2018-11-03-working-with-dates-in-r.html"
## [100] "2018-11-15-working-with-categorical-data-using-forcats.Rmd"
## [101] "2018-11-15-working-with-categorical-data-using-forcats.html"
## [102] "2018-11-27-quick-guide-r-sqlite.Rmd"
## [103] "2018-11-27-quick-guide-r-sqlite.html"
## [104] "2018-12-09-data-wrangling-with-dbplyr.Rmd"
## [105] "2018-12-09-data-wrangling-with-dbplyr.html"
## [106] "2018-12-21-sql-for-data-science.Rmd"
## [107] "2018-12-21-sql-for-data-science.html"
## [108] "2019-01-02-sql-for-data-science-part-2.Rmd"
## [109] "2019-01-02-sql-for-data-science-part-2.html"
## [110] "2019-02-08-introducing-olsrr.Rmd"
## [111] "2019-02-08-introducing-olsrr.html"
## [112] "2019-02-12-introducing-rfm.Rmd"
## [113] "2019-02-12-introducing-rfm.html"
## [114] "2019-02-19-introducing-descriptr.Rmd"
## [115] "2019-02-19-introducing-descriptr.html"
## [116] "2019-02-20-introducing-descriptr.Rmd"
## [117] "2019-02-20-introducing-descriptr.html"
## [118] "2019-02-26-introducing-blorr.Rmd"
## [119] "2019-02-26-introducing-blorr.html"
## [120] "2019-03-05-getting-help-in-r.Rmd"
## [121] "2019-03-05-getting-help-in-r.html"
## [122] "2019-03-12-introducing-rbin.Rmd"
## [123] "2019-03-12-introducing-rbin.html"
## [124] "2019-03-14-introducing-vistributions.Rmd"
## [125] "2019-03-14-introducing-vistributions.html"
## [126] "2019-04-01-shiny-apps.Rmd"
## [127] "2019-04-01-shiny-apps.html"
## [128] "2019-04-11-web-scraping.Rmd"
## [129] "2019-04-11-web-scraping.html"
## [130] "2019-04-13-web-scraping-note.Rmd"
## [131] "2019-04-13-web-scraping-note.html"
## [132] "2019-05-02-mba.Rmd"
## [133] "2019-05-02-mba.html"
## [134] "2019-05-02-mba_files"
## [135] "2019-05-27-regex.Rmd"
## [136] "2019-05-27-regex.html"
## [137] "2019-07-05-pkginfo.Rmd"
## [138] "2019-07-05-pkginfo.html"
## [139] "2019-07-22-customer-segmentation-using-rfm-analysis.Rmd"
## [140] "2019-07-22-customer-segmentation-using-rfm-analysis.html"
## [141] "2019-07-22-customer-segmentation-using-rfm-analysis_files"
## [142] "2019-08-08-working-with-databases-using-r.Rmd"
## [143] "2019-08-08-working-with-databases-using-r.html"
## [144] "2019-08-08-working-with-databases-using-r_files"
## [145] "2019-10-26-command-line-crash-course.Rmd"
## [146] "2019-10-26-command-line-crash-course_files"
## [147] "2020-01-17-nse2r-intro.Rmd"
## [148] "2020-04-17-handling-date-and-time-in-r.Rmd"
## [149] "2020-04-17-handling-date-and-time-in-r_files"
## [150] "airline.dta"
## [151] "airline.sas7bdat"
## [152] "analysis.R"
## [153] "cline"
## [154] "config.yml"
## [155] "employee.sav"
## [156] "examples"
## [157] "hsb2.csv"
## [158] "hsb3.csv"
## [159] "hsb4.csv"
## [160] "imports_blorr.txt"
## [161] "imports_olsrr.txt"
## [162] "mydatabase.db"
## [163] "mypackage"
## [164] "myproject"
## [165] "online-retail.xlsx"
## [166] "options.R"
## [167] "package_names.csv"
## [168] "release.txt"
## [169] "sample.xls"
## [170] "transaction_data.csv"
## [171] "zip_example.zip"Great! Now, how do we specify the options? The additional options of a
command must be stored as a character vector and specified using the args
argument. In the below example, we delete the examples folder we created earlier while decompressing the zip_example.zip file.
system2(command = "rm",
args = c("-r", "examples"))## character(0)In some cases, we might want to redirect the output. Let us say we are writing
message to a file using the echo command. In this cases, we want the output
to be redirected to the release.txt file. The stdout argument can be used to redirect output to a file or the R console. In the below example, we redirect the output to a file.
system2(command = "echo",
args = c("Great Truth"),
stdout = "release.txt")In the next example, we redirect the output to the R console by setting the value of the stdout argument to TRUE. If you are curious, set the value to FALSE and see what happens.
system2(command = "diff",
args = c("imports_olsrr.txt", "imports_blorr.txt"),
stdout = TRUE)## Warning in system2(command = "diff", args = c("imports_olsrr.txt",
## "imports_blorr.txt"), : running command '"diff" imports_olsrr.txt
## imports_blorr.txt' had status 1## [1] "1,4c1,5" "< car " "< checkmate" "< cli" "< clisymbols"
## [6] "---" "> car" "> caret" "> checkmate" "> clisymbols"
## [11] "> cli"
## attr(,"status")
## [1] 1The run() command from the processx package can be used to execute shell commands as well.
Execute Shell Commands in RStudio
In RStudio, commands can be executed from shell scripts by pressing Ctrl + Enter. Instead of sending the command to the R console, it is redirected to
the terminal where it is executed as shown below.
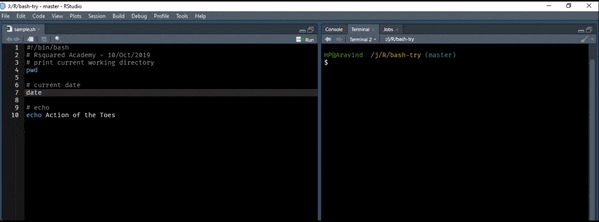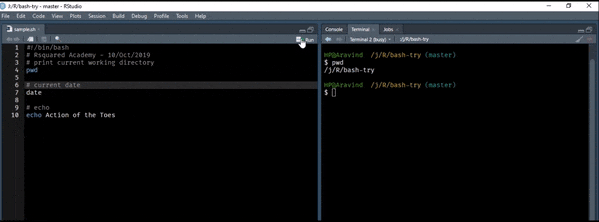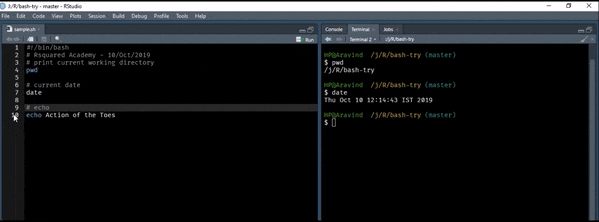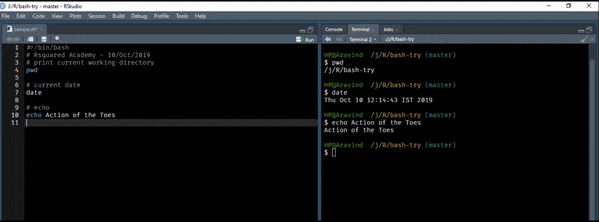
RMarkdown
RMarkdown supports bash, sh and awk. This post was initially created using sh as the underlying operating system is Windows. Later, we used bash after installing the Windows subsystem for Linux. Click here to learn more.
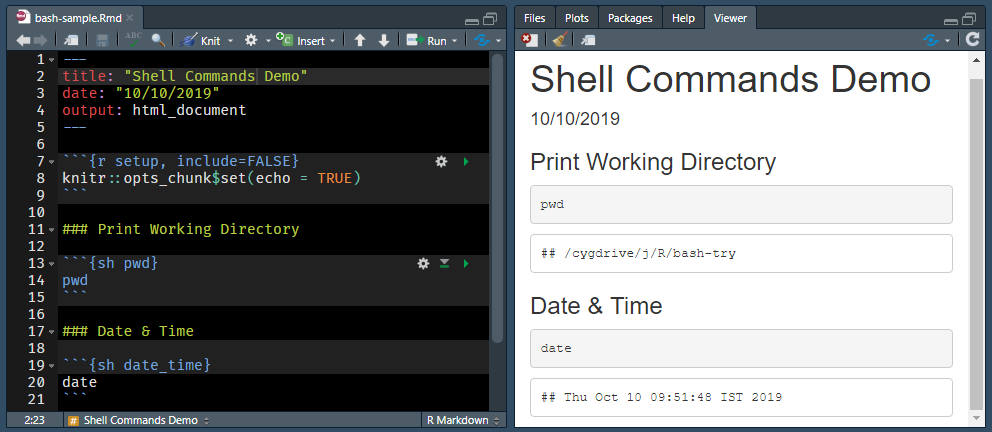
R in the Shell
In this section, we will learn to execute R commands and scripts in the command line using:
R -eRscript -eR CMD BATCH
The -e option allows us to specify R expression(s). R -e will launch R and then execute the code specified within quotes. Use semi-colon to execute multiple expressions as shown below. You will be able to run the below commands only if you are able to launch R from the command line. Windows users need to ensure that R is added to the path environment.
R -e "head(mtcars); tail(mtcars)"##
## R version 4.0.0 (2020-04-24) -- "Arbor Day"
## Copyright (C) 2020 The R Foundation for Statistical Computing
## Platform: x86_64-w64-mingw32/x64 (64-bit)
##
## R is free software and comes with ABSOLUTELY NO WARRANTY.
## You are welcome to redistribute it under certain conditions.
## Type 'license()' or 'licence()' for distribution details.
##
## Natural language support but running in an English locale
##
## R is a collaborative project with many contributors.
## Type 'contributors()' for more information and
## 'citation()' on how to cite R or R packages in publications.
##
## Type 'demo()' for some demos, 'help()' for on-line help, or
## 'help.start()' for an HTML browser interface to help.
## Type 'q()' to quit R.
##
## > head(mtcars); tail(mtcars)
## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
## mpg cyl disp hp drat wt qsec vs am gear carb
## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.7 0 1 5 2
## Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.9 1 1 5 2
## Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.5 0 1 5 4
## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.5 0 1 5 6
## Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.6 0 1 5 8
## Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.6 1 1 4 2
## >
## >Rscript -e will run code without launching R.
Rscript -e "head(mtcars)"## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1We can use Rscript to execute a R script as well. In the below example, we execute the code in analysis.R file.
Rscript analysis.R## mpg cyl disp hp drat wt qsec vs am gear carb
## Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
## Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
## Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
## Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
## Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
## Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1If you are more interested, try the littler package as well.
What we have not covered…
- shell scripting
- editing files
- file permissions
- user information
- pipes
- awk
- sed
Summary
- Shell is a text based application for viewing, handling and manipulating files
- It is also known by the following names
- CLI (Command Line Inteface)
- Terminal
- Bash (Bourne Again Shell)
- Use
system2()andprocessx::run()in R to execute shell commands - Use
Rscript -eorR -eto execute R scripts from the command line - RStudio includes a Terminal (from version 1.1.383)
- Execute commands from shell script in RStudio using Ctrl + Enter
- RMarkdown supports
bash,shandawk
References & Further Readings
- https://tldr.sh/
- https://blog.rstudio.com/2017/08/11/rstudio-v1-1-preview-terminal/
- https://support.rstudio.com/hc/en-us/articles/115010737148-Using-the-RStudio-Terminal
- https://bookdown.org/yihui/rmarkdown/language-engines.html#shell-scripts
- https://github.com/kirillseva/tldrrr
- https://fs.r-lib.org/articles/function-comparisons.html
- https://livefreeordichotomize.com/2017/09/28/r-release-names/
- https://stackoverflow.com/questions/13478375/is-there-any-authoritative-documentation-on-r-release-nicknames
- http://www2.imm.dtu.dk/courses/02333/cygwin_tutorial/index.html
- https://tutorial.djangogirls.org/en/intro_to_command_line/
- https://ss64.com/
- https://www.learnenough.com/command-line-tutorial
Feedback
If you see mistakes or want to suggest changes, please create an issue on the source repository or reach out to us at support@rsquaredacademy.com.


